Software

BATMAN
BATMAN - Bayesian Inference of Activation of TCR by Mutant Antigens:
python package for predicting TCR activation by mutant peptides
based on their distances to the TCR's index peptide. BATMAN can be
used for within-TCR and cross-TCR predictions and in active learning
mode to design TCR activation assays.

copepodTCR
copepodTCR - COmbinatorial PEptide POoling Design for TCR
specificity: python package and webtool for the design of
combinatorial peptide pooling schemes for TCR speficity assays.
CopepodTCR guides the user through all stages of the experimental
design and interpretation, including pooling schemes, peptide mixing
layouts and results evaluation.
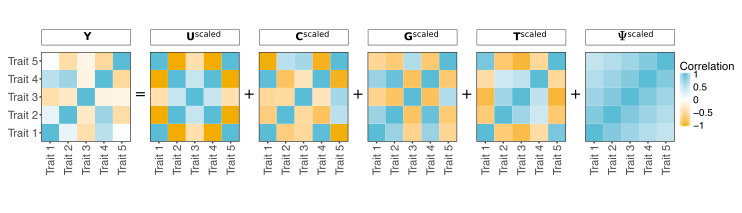
PhenotypeSimulator
R/CRAN framework for simulating genotype to phenotype relationships. It can
model multiple traits with multiple underlying genetic loci as well as complex
covariate and observational noise structure. This package has been designed
to work with many common genetic tools both for input and
output.
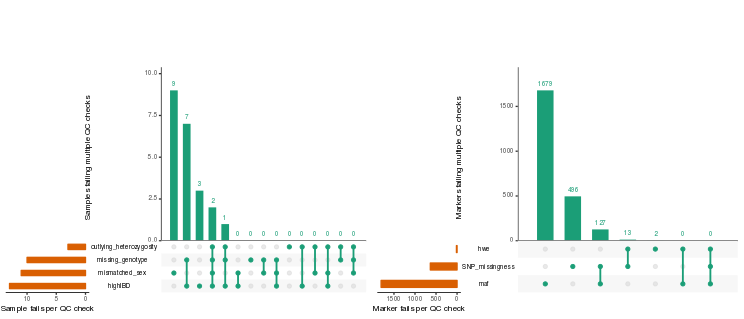
plinkQC
R/CRAN package for common genotype quality control steps. It provides
an easily applicable workflow from per-individual and per-marker quality
control to the generation of the new, quality-controlled dataset.
The outcomes of each quality control step are summarised in a quality
control report.
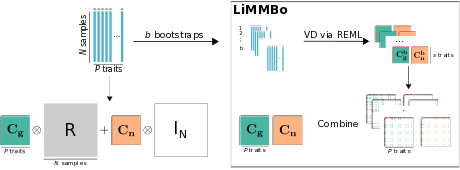
LiMMBo
Python module for computationally efficient joint genetic analysis of
high-dimensional phenotypes, using linear mixed models with bootstrapping.